smooth.optimization package¶
Subpackages¶
Submodules¶
Run Optimization¶
This is the core of the genetic algorithm (GA) used for optimization. It uses the NSGA-II algorithm for multi-objective optimization of smooth components.
How to use¶
To use, call run_optimization with a configuration dictionary and your smooth model.
You will receive a list of Individual in return. These individuals are
pareto-optimal in regard to the given objective functions (limited to two functions).
An example configuration can be seen in run_optimization_example in the examples directory.
Objective functions¶
You may specify your custom objective functions for optimization. These should be lambdas that take the result from run_smooth and return a value. Keep in mind that this algorithm always tries to maximize. In order to minimize a value, return the negative value.
Example 1: maximize power_max of the first component:
lambda x: x[0].power_max
Example 2: minimize the annual costs:
lambda x: -sum([component.results['annuity_total'] for component in x])
Result¶
After the given number of generations or aborting, the result is printed to the terminal. All individuals currently on the pareto front are returned in a list. Their values member contain the component attribute values in the order given by the attribute_variation dictionary from the optimization params. In addition, when SAVE_ALL_SMOOTH_RESULTS is set to True, the smooth_result member of each individual contains the value returned by run_smooth.
Warning
Using SAVE_ALL_SMOOTH_RESULTS and writing the result to a file will generally lead to a large file size.
Implementation¶
Like any GA, this implementation simulates a population which converges to an optimal solution over multiple generations. As there are multiple objectives, the solution takes the form of a pareto-front, where no solution is dominated by another while maintaining distance to each other. We take care to compute each individual configuration only once. The normal phases of a GA still apply:
- selection
- crossover
- mutation
Population initialisation¶
At the start, a population is generated.
The size of the population must be declared (population_size).
Each component attribute to be varied in the smooth_model corresponds
to a gene in an individual. The genes are initialized randomly with a uniform
distribution between the minimum and maximum value of its component attribute.
These values may adhere to a step size (val_step in AttributeVariation).
Selection¶
We compute the fitness of all individuals in parallel. You must set n_core to specify how many threads should be active at the same time. This can be either a number or ‘max’ to use all virtual cores on your machine. The fitness evaluation follows these steps:
- change your smooth model according to the individual’s component attribute values
- run smooth
- on success, compute the objective functions using the smooth result. These are the fitness values. On failure, print the error
- update the master individual on the main thread with the fitness values
- update the reference in the dictionary containing all evaluated individuals
After all individuals in the current generation have been evaluated, they are sorted into tiers by NSGA-II fast non-dominated sorting algorithm. Only individuals on the pareto front are retained, depending on their distance to their neighbors. The parent individuals stay in the population, so they can appear in the pareto front again.
Crossover¶
These individuals form the base of the next generation, they are parents. For each child in the next generation, genes from two randomly selected parents are taken (uniform crossover of independent genes).
Mutation¶
After crossover, each child has a random number of genes mutated. The mutated value is around the original value, taken from a normal distribution. Special care must be taken to stay within the component atrribute’s range and to adhere to a strict step size.
After crossover and mutation, we check that this individual’s gene sequence has not been encountered before (as this would not lead to new information and waste computing time). Only then is it admitted into the new generation.
Special cases¶
We impose an upper limit of 1000 * population_size on the number of tries to find new children. This counter is reset for each generation. If it is exceeded and no new gene sequences have been found, the algorithm aborts and returns the current result.
In case no individuals have a valid smooth result, an entirely new population is generated. No plot will be shown. If only one individual is valid, the population is filled up with random individuals.
Gradient ascent¶
The solutions of the GA are pareto-optimal, but may not be at a local optimum. Although new configurations to be evaluated are searched near the current ones, it is not guaranteed to find slight improvements. This is especially true if there are many dimensions to search and the change is in only one dimension. The chance to happen upon this single improvement is in inverse proportion to the number of attribute variations.
Therefore, the post_processing option exists to follow the fitness gradient for each solution after the GA has finished. We assume that each attribute is independent of each other. All solutions improve the same attribute at the same time. The number of fitness evaluations may exceed the population_size, however, the maximum number of cores used stays the same as before.
To find the local optimum of a single attribute of a solution, we first have to find the gradient. This is done by going one val_step in positive and negative direction. These new children are then evaluated. Depending on the domination, the gradient may be +val_step, -val_step or 0 (parent is optimal). Then, this gradient is followed until the child shows no improvement. The population may be topped up with multiples of val_step to better utilize all cores and speed up the gradient ascent. After all solutions have found their optimum for this attribute, the next attribute is varied.
Plotting¶
To visualize the current progress, you can set the plot_progress simulation parameter to True. This will show the current pareto front in a pyplot window. You can mouse over the points to show the configuration and objective values. To keep the computation running in the background (non-blocking plots) while listening for user events, the plotting runs in its own process.
On initialisation, a one-directional pipe is established to send data from the main computation to the plotting process. The process is started right at the end of the initialisation. It needs the attribute variations and objective names for hover info and axes labels. It also generates a multiprocessing event which checks if the process shall be stopped.
In the main loop of the process, the pipe is checked for any new data. This incorporates a timeout to avoid high processor usage. If new data is available, the old plot is cleared (along with any annotations, labels and titles) and redrawn from scratch. In any case, the window listens for a short time for user input events like mouseover. Window close is a special event which stops the process, but not the computation (as this runs in the separate main process).
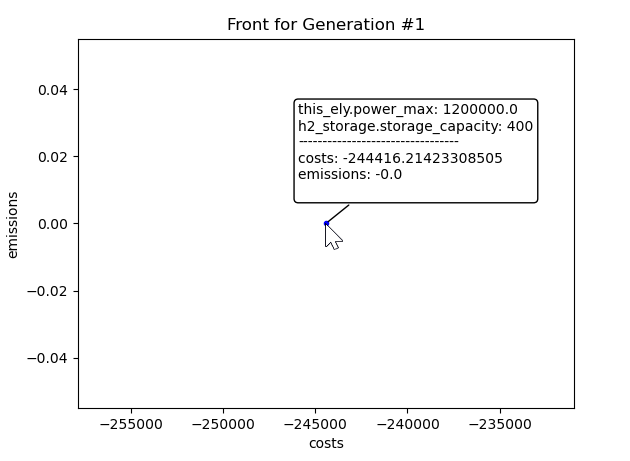
When hovering with the mouse pointer over a point in the pareto front,
an annotation is built with the info of the Individual.
The annotation is removed when leaving the point. A simple example
of how this looks is illustrated in Figure 1. In this example,
after the first generation there is one optimal energy system
found which costs 244,416.21 EUR and produces 0 emissions.
Sending None through the pipe makes the process show the plot until the user closes it. This blocks the process, so no new data is received, but user events are still processed.
-
class
smooth.optimization.run_optimization.AttributeVariation(iterable=(), **kwargs)¶ Bases:
objectClass that contains all information about an attribute that is varied by the genetic algorithm
Parameters: - comp_name (string) – name of component that gets varied
- comp_attribute (string) – component attribute that gets varied
- val_min (number) – minimum value of component attribute
- val_max (number) – maximum value of component attribute (inclusive)
- val_step (number, optional) – step size of component attribute
Variables: num_steps – number of steps if val_step is set and not zero
Raises: AssertionError when any non-optional parameter is missing or val_step is negative
-
class
smooth.optimization.run_optimization.Individual(values)¶ Bases:
objectClass for individuals evaluated by the genetic algorithm
Parameters: values (list) – attribute values (individual configuration)
Variables: - values – given values
- fitness – fitness values depending on objective functions
- smooth_result – result from run_smooth
-
class
IndividualIterator(individual)¶ Bases:
objectClass to iterate over gene values.
-
fitness= None¶
-
smooth_result= None¶
-
values= None¶
-
smooth.optimization.run_optimization.crossover(parent1, parent2)¶ - Uniform crossover between two parents
- Selects random (independent) genes from one parent or the other
Parameters: - parent1 (
Individual) – First parent - parent2 (
Individual) – Second parent
Returns: Crossover between parents
Return type:
-
smooth.optimization.run_optimization.mutate(parent, attribute_variation)¶ Mutate a random number of parent genes around original value, within variation
Parameters: - parent (
Individual) – parent individual - attribute_variation (list of
AttributeVariation) – AV for all genes in parent
Returns: child with some parent genes randomly mutated
Return type: - parent (
-
smooth.optimization.run_optimization.fitness_function(index, individual, model, attribute_variation, dill_objectives, ignore_zero=False, save_results=False)¶ - Compute fitness for one individual
- Called async: copies of individual and model given
Parameters: - index (int) – index within population
- individual (
Individual) – individual to evaluate - model (dict) – smooth model
- attribute_variation (list of
AttributeVariation) – attribute variations - dill_objectives (tuple of lambda-functions pickled with dill) – objective functions
- ignore_zero (boolean) – ignore components with an attribute value of zero
- save_results (boolean) – save smooth result in individual?
Returns: index, modified individual with fitness (None if failed) and smooth_result (none if not save_results) set
Return type: tuple(int,
Individual)
-
class
smooth.optimization.run_optimization.PlottingProcess¶ Bases:
multiprocessing.context.ProcessProcess for plotting the intermediate results
Data is sent through (onedirectional) pipe. It should be a dictionary containing “values” (array of
Individual) and “gen” (current generation number, displayed in title). Send None to stop listening for new data and block the Process by showing the plot. After the user closes the plot, the process returns and can be joined.Parameters: - pipe (multiprocessing pipe) – data transfer channel
- attribute_variation (list of
AttributeVariation) – AV ofOptimization - objective_names (list of strings) – descriptive names of
Optimizationobjectives
Variables: - exit_flag – Multiprocessing event signalling process should be stopped
- fig – figure for plotting
- ax – current graphic axis for plotting
- points – plotted results or None
- annot – current annotation or None
-
main()¶ Main plotting thread
Loops while exit_flag is not set and user has not closed window. Checks periodically for new data to be displayed.
-
handle_close(event)¶ Called when user closes window
Signal main loop that process should be stopped.
-
hover(event)¶ Called when user hovers over plot.
Checks if user hovers over point. If so, delete old annotation and create new one with relevant info from all Indivdiuals corresponding to this point. If user does not hover over point, remove annotation, if any.
-
class
smooth.optimization.run_optimization.Optimization(iterable=(), **kwargs)¶ Bases:
objectMain optimization class to save GA parameters
Parameters: - n_core (int or 'max') – number of threads to use. May be ‘max’ to use all (virtual) cores
- n_generation (int) – number of generation to run
- population_size (int) – number of new children per generation. The actual size of the population may be higher - however, each individual is only evaluated once
- attribute_variation (list of dicts, see
AttributeVariation) – attribute variation information that will be used by the GA - model (dict) – smooth model
- objectives (2-tuple of lambda functions) – multi-objectives to optimize. These functions take the result from run_smooth and return a float. Positive sign maximizes, negative sign minimizes. Defaults to minimizing annual costs and emissions
- objective_names (2-tuple of strings, optional) – descriptive names for optimization functions. Defaults to (‘costs’, ‘emissions’)
- post_processing (boolean, optional) – improve GA solution with gradient ascent. Defaults to False
- plot_progress (boolean, optional) – plot current pareto front. Defaults to False
- ignore_zero (boolean, optional) – ignore components with an attribute value of zero. Defaults to False
- save_intermediate_results (boolean, optional) – write intermediate results to pickle file. Only the two most recent results are saved. Defaults to False
- SAVE_ALL_SMOOTH_RESULTS (boolean, optional) – save return value of run_smooth for all evaluated individuals. Warning! When writing the result to file, this may greatly increase the file size. Defaults to False
Variables: - population – current individuals
- evaluated – keeps track of evaluated individuals to avoid double computation
- ax – current figure handle for plotting
Raises: AttributeError or AssertionError when required argument is missing or wrong
-
err_callback(err_msg)¶ Async error callback
Parameters: err_msg (string) – error message to print
-
set_fitness(result)¶ Async success callback Update master individual in population and evaluated dictionary
Parameters: result (tuple(index, Individual)) – result from fitness_function
-
compute_fitness()¶ Compute fitness of every individual in population with n_core worker threads. Remove invalid individuals from population
-
save_intermediate_result(result)¶ Dump result into pickle file in current working directory. Same content as smooth.save_results. The naming schema follows date-time-intermediate_result.pickle. Removes second-to-last pickle file from same run.
Parameters: result (list of Individual) – the current results to be saved
-
gradient_ascent(result)¶ Try to fine-tune result(s) with gradient ascent
Attributes are assumed to be independent and varied separately. Solutions with the same fitness are ignored.
Parameters: result (list of Individual) – result from GAReturns: improved result Return type: list of Individual
-
run()¶ Main GA function
Returns: pareto-optimal configurations Return type: list of Individual
-
smooth.optimization.run_optimization.run_optimization(opt_config, _model)¶ Entry point for genetic algorithm
Parameters: - opt_config (dict) – Optimization parameters.
May have separate ga_params dictionary or define parameters directly.
See
Optimization. - _model (dict or list (legacy)) – smooth model
Returns: pareto-optimal configurations
Return type: list of
Individual- opt_config (dict) – Optimization parameters.
May have separate ga_params dictionary or define parameters directly.
See